3 hours or so for excellent results, there of couse is another method called qRT-PCR which also quantifies contaminants and thus eliminates electrophoresis in cases, but this will require operations to be run with different primers for each contaminant, from what I know, and this could complicate things a lot. You may want to talk to qRT-PCR manufacturers to get some advice on the technique, since I haven't used it I cannot offer any advice.ScholasticSpastic wrote:How quantitative is this? If it's +/- 10% or better, it's probably adequate for what these fellas want. Considered similar ideas but didn't know whether we could obtain quantitative results.GenesForLife wrote:well, 16S RNA can be used to identify a species, and you can have fluorescent probes et cetera for that, there is a technique called Rt-PCR which amplifies DNA prepared from the RNA of the cells from the culture. You can then carry out electrophoresis and carry out Southern Blotting, with the probe included, just to see if there are 16S rRNA sequences from other species showing up too.ScholasticSpastic wrote:GenesForLife wrote:if they are talking about other algal contaminants, the idea is PCR + species specific probe hybridization.I'ma learn what that means now!
1) Identify species marker for the non-contaminant organism.
2) Use Rt-PCR to amplify DNA from culture.
3) Separate by Agarose Gel Electrophoresis.
4) See if all the bands hybridize with the fluorescent probe or if just a few will.
5) If some bands go unhybridized they indicate contaminants.

I am going to check later to see if I can design primers that amplify a wide range of 16S rRNA primers from algae, btw, PCR + probe hybridization is a highly senstive assay and can detect contaminants from small samples of rRNA/DNA.
Please note, for evaluating rRNA samples you need to use Rt PCR (where Reverse transcriptase converts 16S rRNA to DNA, which then is amplified by PCR)
Ignoring specifics, and given novice technicians, what's a reasonable expectation for sample turnaround time? Will we be able to discriminate between contributions from contaminant bacteria and unwanted algae without too much further effort?
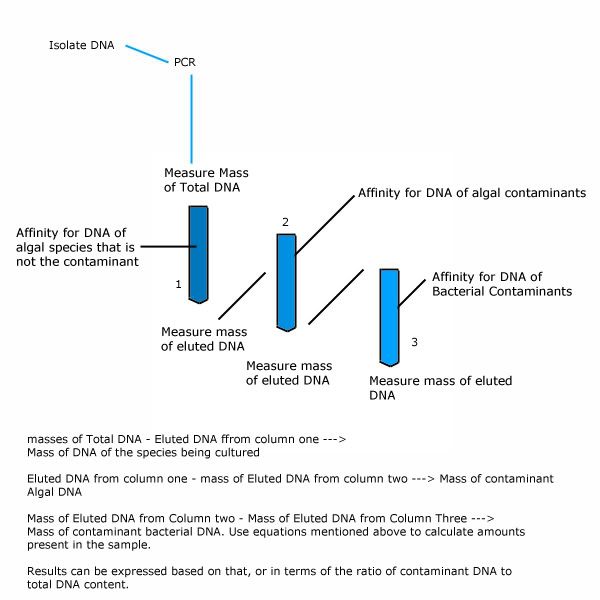
Quantification could be done after electrophoresis, the gel can be melted, the DNA can be reisolated and used to calculate the amount of original contaminant DNA that was amplified since we know how many doublings have taken place to arrive at the amount recovered after electrophoresis. This is done by means of a simple assay using a spectrophotometer or colorimeter.
The glitch here is to identify what number of 16S rRNA copies have been produced by amplification based on the DNA concentration, if that challenge can be worked out then I see no reason why this cannot make a very good solution for your needs.
To distinguish between bacteria and algal contaminants too, the PCR protocol will require some changes too, I will get back to you on this once I have it sorted in my head.